12 Oct 2021
HKU Biologists build a megabase-sized chromosome from scratch Further exploring the potential of worm artificial chromosomes to improve gene therapy
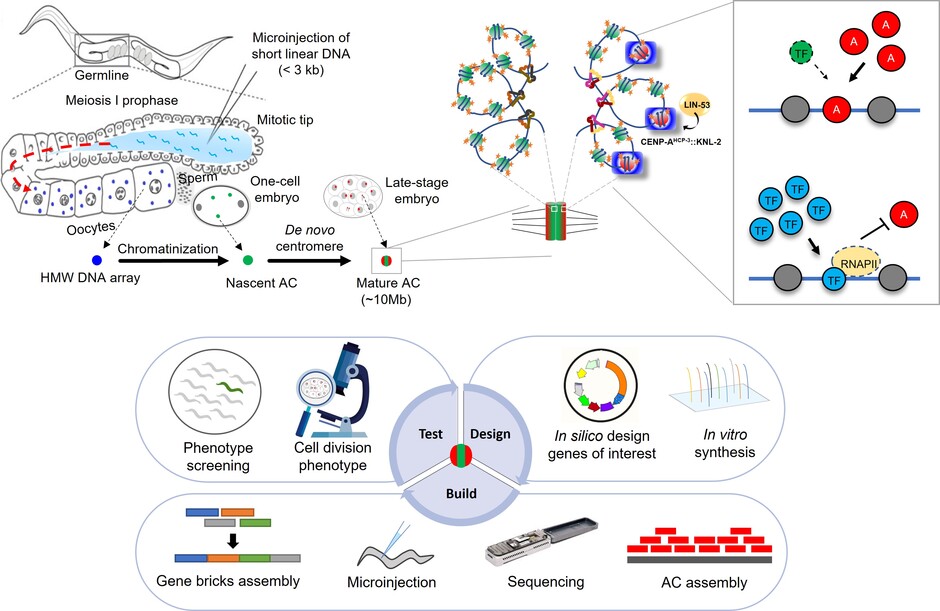
Image credit: Upper panel: Nucleic Acids Research, Lower panel: by Lin Zhongyang.
A research team led by Dr Karen Wing Yee YUEN, Associate Professor from the School of Biological Sciences at The University of Hong Kong (HKU), revealed the mechanism of artificial chromosome (AC) formation in the embryos of the model organism Caenorhabditis elegans, a 1-mm long, transparent nematode.
The findings were recently published as two back-to-back papers in the influential scientific journal Nucleic Acids Research. These studies have provided insights into the mechanisms of DNA assembly, new centromere formation and have facilitated the engineering of ACs for cloning and gene therapy. In summary, Dr Karen Yuen and postdoctoral fellow Dr Zhongyang LIN have discovered the cellular proteins in vivo (inside a living organism) used for processing foreign, naked DNA fragments to form a chromatin-packaged artificial chromosome and dissected the molecular mechanisms of how kb-sized foreign DNA can be assembled into an over 10 megabase-sized artificial chromosome in C. elegans.
What are artificial chromosomes, and why do they hold the key to future medicine?
Essentially, our DNA is packaged meticulously by proteins to make up the chromatin. If DNA were like a thread, these proteins are the spools that the DNA thread winds around to keep itself organised and neat inside of a microscopic cell. However, what will happen when a foreign, naked DNA thread with no spool is introduced into the environment? Interestingly, the cell is equipped to supply this new thread with its own self-made spools, enabling this naked DNA thread to be stably maintained in the cellular environment as part of the cell’s new repertoire. We call this process artificial chromosome (AC) formation.
Amongst the useful applications of artificial chromosomes, one of the most exciting prospects is gene therapy. For example, the fatal, chronic lung disease cystic fibrosis (CF) is caused by a mutation in the CFTR gene and is currently an incurable disease. Scientists have been studying the use of bacterial and yeast artificial chromosomes (BACs and YACs) as a vector or carrier to express the normal, functional CFTR gene and overcome the defective CFTR expression in patient cells.
As in life, to manipulate something, we must first understand it. In order to engineer artificial chromosomes, we must first learn how they are formed and maintained.
How new chromosomes are formed and maintained, and the importance of the centromere
Almost two trillion cells divide every day in an average human body. This means that two trillion cells have to make a perfect copy of themselves every time. The cost of cell division that comes short of flawlessness is undoubtedly humankind’s worst enemy yet: cancer, in which many are characterised by chromosome instability. One important player in ensuring the faithful inheritance of our chromosomes during cell division is the centromere.
The centromere is a specialised region on each chromosome that connects the chromosome to spindle microtubules to orchestrate chromosome segregation in every cell division. In some cancer cells, centromeres may become inactivated or lost from chromosomal rearrangements and bypass chromosome loss by forming a new centromere on a random, ectopic region. So far, not much is known about new centromere (neocentromere) formation, despite it being implicated in chromosome instability and a driver of tumorigenesis. This is because neocentromere formation is notoriously difficult to study since the process of neocentromere establishment has been challenging to observe, as they are only ascertained when developmental disorders or cancer arise and genomic analyses are performed. In other words, new centromeres are often detected long after their formation and stabilisation.
To conduct studies on centromere formation, Dr Yuen’s team utilised a straightforward, direct method: microinjection, developed almost 30 years ago. In other species, such as humans, foreign DNA is recognised and excluded mostly and hence not propagated into future generations as a self-defence mechanism. Surprisingly, C. elegans is one of the rare species that allows foreign DNA, completely devoid of any C. elegans DNA sequence, to fuse into a big, megabase-sized artificial chromosome. Simply put, while C. elegans can build an artificial chromosome with no native sequence requirement, the same cannot be said about other artificial chromosomes from other species like human (HAC), which require some human DNA sequences in order to build and propagate as an artificial chromosome.
To further study this unique characteristic, the team has developed an in vivo fluorescent system to visualise the artificial chromosomes in real-time. Dr Yuen’s lab uses this artificial chromosome segregation assay as a functional readout for de novo (from the beginning) centromere formation to investigate factors that affect de novo centromere establishment. Dr Lin identified the histone chaperone RbAp46/48LIN-53 and acetyltransferase HAT-1 as essential for centromere formation. Here, C. elegans acts as a robust model due to its transparent embryos that facilitate imaging, but also its rare nature to efficiently induce de novo centromere formation and segregate the new artificial chromosomes faithfully within a few cell cycles in the embryos.
The current studies of artificial chromosomes provide novel insights into the chromosomal processes required for de novo centromere formation and chromosome maintenance. Dr Yuen’s team revealed the in vivo biological processes necessary for exogenous DNA to become a stably propagating artificial chromosome and unravel the hierarchy in assembling a de novo centromere from foreign DNA fragments. To further dissect the unique characteristic of C. elegans in adopting foreign DNA sequences, Dr Yuen’s team also explored if this phenomenon is bound by rules, i.e. manipulating the DNA sequence composition, complexity and length to observe the preferences for building an artificial chromosome in C. elegans.
Using these approaches, we are now able to observe and systematically compare how a new centromere is established on an artificial chromosome and also how a pre-existing centromere is maintained on the C. elegans endogenous chromosomes.
Can artificial chromosomes in worms be the answer to gene therapy?
Finally, how are worms relevant to humans? While the order of how genes are arranged on a chromosome can be consistent across different closely related species, centromere repositioning also occurs throughout evolution. Therefore, the process of new centromere formation may act as a driver of diseases but also serves as a marker of evolution. Furthermore, the results of these studies could help advance the synthetic biology field by exploring how some characteristics can be designed to optimise the establishment of an artificial chromosome by improving the efficiency of de novo centromere formation through accurate segregation to improve the applications of ACs as large-capacity, faithful vectors for cloning and gene therapies.
The latest findings can be accessed from the following link.
And the complementary journal paper can be accessed from here.